Module tf.dataset.nodemaps
Node maps
When different versions of a TF dataset exist, it makes sense to map the nodes from the older version to nodes of the newer version.
If we have annotations on the older version (possibly created with considerable man-machine labour), we can use such node mappings to transfer the annotations to the newer version without redoing all the work.
Mapping the nodes consists of two stages:
- slot mapping This is very dependent on what happened between versions.
- node mappings per node type These can be generically constructed once we have a slot mapping.
This module contains the function makeVersionMapping which is a function to furnish node mappings
for node types given a slot mapping exists.
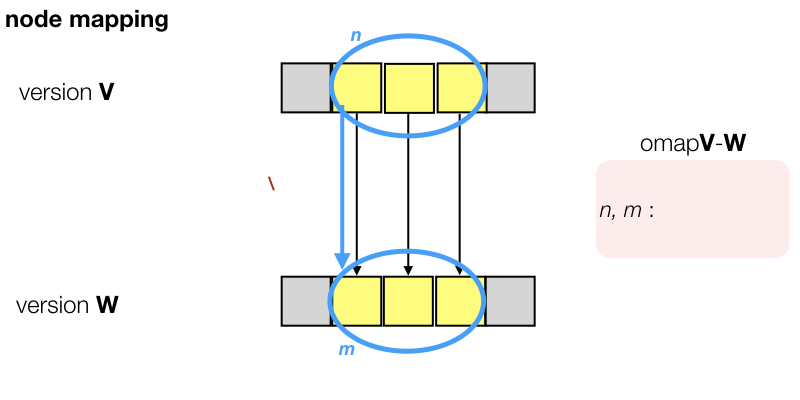
Nodes in general
The basic idea we use for the general case is that that nodes are linked to slots.
In TF, the standard oslots edge feature lists for each non-slot node
the slots it is linked to.
Combining the just created slot mappings between versions and the oslots feature,
we can extend the slot mapping into a general node mapping.
In order to map a node n in version V, we look at its slots s, use the already established slot mapping to map these to slots t in version W, and collect the nodes m in version W that are linked to those t. They are good candidates for the mapping.
Refinements
When we try to match nodes across versions, based on slot containment, we also respect
their otype. So we will not try to match a clause to a phrase.
We make implicit use of the fact that for most otype, the members
contain disjoint slots.
Multiple candidates
Of course, things are not always as neat as in the diagram. Textual objects may have split, or shifted, or collapsed. In general we find 0 or more candidates. Even if we find exactly one candidate, it does not have to be a perfect match. A typical situation is this:
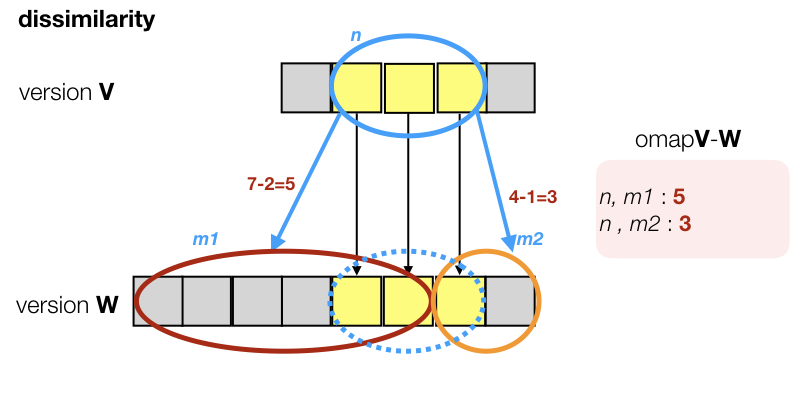
We do not find a node m\in W that occupies the mapped slots exactly. Instead, we find that the target area is split between two candidates who also reach outside the target area.
In such cases, we make edges to all such candidates, but we add a dissimilarity measure. If m is the collection of slots, mapped from n, and m_1 is a candidate for n, meaning m_1 has overlap with m, then the dissimilarity of m_1 is defined as:
|m_1\cup m| - |m_1\cap m|
In words: the number of slots in the union of m_1 and m minus the number of slots in their intersection.
In other words: m_1 gets a penalty for
- each slot s\in m_1 that is not in the mapped slots m;
- each mapped slot t\in m that is not in m_1.
If a candidate occupies exactly the mapped slots, the dissimilarity is 0. If there is only one such candidate of the right type, the case is completely clear, and we do not add a dissimilarity value to the edge.
If there are more candidates, all of them will get an edge, and those edges will contain the dissimilarity value, even if that value is 0.
Diagnostic labels
We report the success in establishing the match between non-slot nodes. We do so per node type, and for each node type we list a few statistics, both in absolute numbers and in percentage of the total amount of nodes of that type in the source version.
We count the nodes that fall in each of the following cases. The list of cases is ordered by decreasing success of the mapping.
bunique, perfect: there is only one match for the mapping and it is a perfect one in terms of slots linked to it;dmultiple, one perfect: there are multiple matches, but at least one is perfect; this occurs typically if nodes of a type are linked to nested and overlapping sequences of slots;cunique, imperfect: there is only one match, but it is not perfect; this indicates that some boundary reorganization has happened between the two versions, and that some slots of the source node have been cut off in the target node; yet the fact that the source node and the target node correspond is clear;fmultiple, cleanly composed: in this case the source node corresponds to a bunch of matches, which together cleanly cover the mapped slots of the source node; in other words: the original node has been split in several parts;emultiple, non-perfect: all remaining cases where there are matches; these situations can be the result of more intrusive changes; if it turns out to be a small set they do require closer inspection;anot mapped: these are nodes for which no match could be found.
An involved example of creating node mappings between versions (not using this code) is versionMappings.ipynb.
A simpler example, using this code is map.ipynb.
Usage
from tf.dataset import Versions
V = Versions(api, va, vb, slotMap)
V.makeVersionMapping()
Classes
class Versions (api, va, vb, silent='auto', slotMap=None)-
Expand source code Browse git
class Versions: def __init__(self, api, va, vb, silent=SILENT_D, slotMap=None): """Map the nodes of a node type between two versions of a TF dataset. There are two scenarios in which you can use this object: **Extend a slot map to a node mapping**. If this object is initialized with a `slotMap`, it can extend that mapping to a complete node mapping and save the mapping as an `f"omap@{va}-{vb}"` edge feature in the dataset version `vb`. In this use case, the APIs for both `va` and `vb` should be full TF APIs, in particular, they should provide the `tf.core.locality` API. **Use an existing node map to upgrade features from one version to another**. In this use case, you do not have to pass a `slotMap`, but in the `vb` dataset there should be an `omap@a-b` feature, i.e. a node map from the nodes of `va` to those of `vb`. The APIs for both `va` and `vb` may be partial TF APIs, which means that they only have to provide the `tf.core.nodefeature.NodeFeatures` (`F`) and `tf.core.edgefeature.EdgeFeatures` (`E`) APIs. Parameters ---------- api: dict TF API objects to several versions of a dataset, keyed by version label va: string version label of the version whose nodes are the source of the mapping vb: string version label of the version whose nodes are the target of the mapping slotMap: dict, optional None The actual mapping between slots of the old version and the new version silent: string, optional tf.core.timestamp.SILENT_D See `tf.core.timestamp.Timestamp` """ self.silent = silentConvert(silent) self.api = api self.va = va self.vb = vb self.edge = slotMap self.good = True for v in (va, vb): if v not in api: sys.stderr.write( f"No TF API for version {va} in the `api` parameter.\n" ) self.good = False if not self.good: return # self.La = api[va].L if slotMap is not None: self.Lb = api[vb].L self.Ea = api[va].E self.Esa = api[va].Es self.Eb = api[vb].E self.Esb = api[vb].Es self.Fa = api[va].F self.Fsa = api[va].Fs self.Fb = api[vb].F self.Fsb = api[vb].Fs self.TFa = api[va].TF self.TFb = api[vb].TF self.info = self.TFb.info self.warning = self.TFb.warning self.error = self.TFb.error self.diagnosis = {} def makeNodeMapping(self, nodeType): edge = self.edge if edge is None: self.error("Cannot make node mapping if no slot mapping is given") return False va = self.va vb = self.vb Lb = self.Lb Ea = self.Ea Eb = self.Eb Fa = self.Fa Lub = Lb.u Eosa = Ea.oslots.s Eosb = Eb.oslots.s otypesa = Fa.otype.s self.caption(2, f"Mapping {nodeType} nodes {va} ==> {vb}") diag = {} self.caption(0, f"Extending slot mapping {va} ==> {va} for {nodeType} nodes") for n in otypesa(nodeType): slots = Eosa(n) mappedSlots = set( chain.from_iterable(set(edge[s]) for s in slots if s in edge) ) mappedNodes = set( chain.from_iterable(set(Lub(s, nodeType)) for s in mappedSlots) ) result = {} nMs = len(mappedNodes) if nMs == 0: diag[n] = "a" elif nMs >= 1: theseMSlots = {} for m in mappedNodes: mSlots = set(Eosb(m)) dis = len(mappedSlots | mSlots) - len(mappedSlots & mSlots) result[m] = dis theseMSlots[m] = mSlots # we wait further case analysis before we put these counterparts of n into the edge if nMs == 1: m = list(mappedNodes)[0] dis = result[m] if dis == 0: diag[n] = "b" edge[n] = {m: None} # this is the most freqent case, hence an optimization: no dis value. # all other cases require the dis value to be passed on, even if 0 else: diag[n] = "c" edge[n] = {m: dis} else: edge[n] = result dis = min(result.values()) if dis == 0: diag[n] = "d" else: allMSlots = set( chain.from_iterable( set(theseMSlots[m]) for m in mappedNodes ) ) composed = allMSlots == mappedSlots and sum( result.values() ) == len(mappedSlots) * (len(mappedNodes) - 1) if composed: diag[n] = "f" else: diag[n] = "e" self.diagnosis[nodeType] = diag self.caption(0, "\tDone") def exploreNodeMapping(self, nodeType): va = self.va vb = self.vb diagnosis = self.diagnosis self.caption(4, "Statistics for {} ==> {} ({})".format(va, vb, nodeType)) diag = diagnosis[nodeType] total = len(diag) if total == 0: return reasons = collections.Counter() for (n, dia) in diag.items(): reasons[dia] += 1 self.caption(-1, "\t{:<30} : {:6.2f}% {:>7}x".format("TOTAL", 100, total)) for stat in STAT_LABELS: statLabel = STAT_LABELS[stat] amount = reasons[stat] if amount == 0: continue perc = 100 * amount / total self.caption( -1, "\t{:<30} : {:6.2f}% {:>7}x".format(statLabel, perc, amount) ) def getDiagnosis(self, node=None, label=None): """Show the diagnosis of a mapping. You can get various amounts of information about this: * A dictionary that maps all nodes of a given type to a diagnostic label * A list of nodes of a node type that have a given diagnostic label * The label of a given node Parameters ---------- node: string | integer, optional None The node or node type that you want to diagnose. If a string, it is a node type, and you get information about the nodes of that node type. If an int, it is a node, and you get information about that node. If `None`, you get information about all nodes. label: char, optional None If given, it is a diagnostic label (see `tf.dataset.nodemaps`), and the result is dependent on the value passed to *node*: if that is a single node, a boolean is returned telling whether that node has the given label; if that is a node type, a tuple is returned of all nodes in that node type that have this label; if that is None, a tuple is returned of all nodes with that label. If not given, the result depends again on the value passed to *node*: if that is a single node, you get its diagnostic label; if that is a node type, you get a dict, keyed by node, and valued by diagnostic label, for all nodes in the node type; if that is None, you get a dictionary keyed by node, and valued by diagnostic label but now for all nodes. """ diagnosis = self.diagnosis Fa = self.Fa otypesa = Fa.otype.s if label is None: return ( dict(chain.from_iterable(x.items() for x in diagnosis.values())) if node is None else diagnosis.get(node, {}) if type(node) is str else diagnosis.get(otypesa(node), {}).get(node, None) if type(node) is int else None ) else: return ( tuple( y[0] for y in chain.from_iterable(x.items() for x in diagnosis.values()) if y[1] == label ) if node is None else tuple( y[0] for y in diagnosis.get(node, {}).items() if y[1] == label ) if type(node) is str else diagnosis.get(otypesa(node), {}).get(node, None) == label if type(node) is int else None ) def legend(self): """Show the labels and descriptions of the diagnosis classes. When the mapping turns out to be not perfect for a node, the result can be categorized according to severity. This function shows that classification. See also `tf.dataset.nodemaps`. """ for (acro, desc) in STAT_LABELS.items(): self.info(f"{acro} = {desc}", tm=False) def omapName(self): va = self.va vb = self.vb return f"omap@{va}-{vb}" def writeMap(self): TFb = self.TFb va = self.va vb = self.vb edge = self.edge silent = self.silent fName = self.omapName() self.caption(4, "Write edge as TF feature {}".format(fName)) edgeFeatures = {fName: edge} metaData = { fName: { "about": "Mapping from the slots of version {} to version {}".format( va, vb ), "encoder": "TF tf.dataset.nodemaps", "valueType": "int", "edgeValues": True, } } TFb.save( nodeFeatures={}, edgeFeatures=edgeFeatures, metaData=metaData, silent=silent, ) def makeVersionMapping(self): Fa = self.Fa self.diagnosis = {} nodeTypes = Fa.otype.all for nodeType in nodeTypes[0:-1]: self.makeNodeMapping(nodeType) self.exploreNodeMapping(nodeType) self.writeMap() def migrateFeatures(self, featureNames, silent=None, location=None): """Migrate features from one version to another based on a node map. If you have a dataset with several features, and if there is a node map between the versions, you can migrate features from the older version to the newer. The naive approach per feature is: For each node in the old version that has a value for the feature, map the node to the new version. That could be several nodes. Then give all those nodes the value found on the node in the old set. The problem with that is this: Suppose node `n` in the old version maps to nodes `p` and `q` in the new version, but not with the same quality. Suppose the value is `v`. Then the feature assignment will assign `v` to nodes `p` and `q`. But there could very well be a later node `m` with value `w` in the old version that also maps to `p`, and with a lower quality. Then later the node `p` in the new version will be assigned the value `w` because of this, and this is sub optimal. So we make sure that for each feature and each node in the new version the value assigned to it is the value of the node in the old version that maps with the highest quality to the new node. Parameters ---------- featureNames: tuple The names of the features to migrate. They may be node features or edge features or both. location: string, optional None If absent, the migrated features will be saved in the newer dataset. Otherwise it is a path where the new features should be saved. silent: string, optional None See `tf.core.timestamp.Timestamp` This will override the `silent` setting that has been used when creating the `Versions` object. """ silent = self.silent if silent is None else silentConvert(silent) omapName = self.omapName() TFa = self.TFa TFb = self.TFb TFb.setSilent(silent) TFb.warning("start migrating") TFb.load(omapName, add=True, silent=silent) Fsa = self.Fsa Esa = self.Esa Esb = self.Esb va = self.va vb = self.vb emap = Esb(omapName).f nodeFeatures = {} edgeFeatures = {} metaData = {} def getBestMatches(n, dest): result = [] mapped = emap(n) if mapped: perfect = [x for x in mapped if x[1] is None] if perfect: return [perfect[0]] nearPerfect = [x for x in mapped if x[1] == 0] if nearPerfect: (m, q) = nearPerfect[0] if m not in dest or m in dest and dest[m][1] is not None: return [nearPerfect[0]] for (m, q) in mapped: # these q cannot be None or 0 if m in dest: (origVal, origQ) = dest[m] higherQuality = origQ is not None and q < origQ if origVal == val: if higherQuality: result.append((m, q)) elif higherQuality: result.append((m, q)) else: result.append((m, q)) return result for featureName in featureNames: isEdge = TFa.features[featureName].isEdge TFb.info(f"Mapping {featureName} ({'edge' if isEdge else 'node'})") featureObj = (Esa if isEdge else Fsa)(featureName) featureInfo = dict(featureObj.items()) metaData[featureName] = featureObj.meta metaData[featureName]["upgraded"] = f"‼️ from version {va} to {vb}" edgeValues = featureObj.doValues if isEdge else False dest = edgeFeatures if isEdge else nodeFeatures dest[featureName] = {} dest = dest[featureName] # we will not only store the feature value in dest, # but also the quality of the mapping that # produced the value. # We only override an existing value # if the quality of the mapping is better. # Moreover, if one mapping has quality 0, # it means that the mapping is perfect regarding the material in both nodes # but there are lesser mappings as well. # In those cases we do not extend the value to those lesser mappings # Later, we strip the qualities again. if isEdge: if edgeValues: for (n, ts) in featureInfo.items(): matched = getBestMatches(n, dest) if matched: matchedTs = {} for (t, val) in ts.items(): for (t, r) in getBestMatches(t, matchedTs): matchedTs[t] = (val, r) for t in matchedTs: matchedTs[t] = matchedTs[t][0] for (m, q) in matched: dest[m] = (matchedTs, q) """ mapped = emap(n) if mapped: mappedts = {} for (t, val) in ts.items(): tmapped = emap(t) if tmapped: for tEntry in tmapped: mappedts[tEntry[0]] = val for entry in mapped: dest[entry[0]] = mappedts """ else: for (n, ts) in featureInfo.items(): matched = getBestMatches(n, dest) if matched: matchedTs = {} for t in ts: for (t, r) in getBestMatches(t, matchedTs): matchedTs[t] = (None, r) matchedTs = set(matchedTs) for (m, q) in matched: dest[m] = (matchedTs, q) """ mapped = emap(n) if mapped: mappedts = set() for t in ts: tmapped = emap(t) if tmapped: for tEntry in tmapped: mappedts.add(tEntry[0]) for entry in mapped: dest[entry[0]] = mappedts """ else: for (n, val) in featureInfo.items(): for (m, q) in getBestMatches(n, dest): dest[m] = (val, q) for m in dest: dest[m] = dest[m][0] TFb.save( nodeFeatures=nodeFeatures, edgeFeatures=edgeFeatures, metaData=metaData, location=location, module=vb, silent=silent, ) TFb.warning("Done") def caption(self, level, heading, good=None, newLine=True, continuation=False): silent = self.silent prefix = "" if good is None else "SUCCES " if good else "FAILURE " reportHeading = "{}{}".format(prefix, heading) emit = ( silent == VERBOSE or silent == AUTO and level in {-1, 2, 3, 4} or silent == TERSE and level in {2, 3} ) if level in {-1, 0}: # non-heading message decoration = "" if continuation else "| " formattedString = """{}{}""".format(decoration, reportHeading) elif level == 1: # pipeline level formattedString = """ ##{}## # {} # # {} # # {} # ##{}## """.format( "#" * 90, " " * 90, "{:<90}".format(reportHeading), " " * 90, "#" * 90, ) elif level == 2: # repo level formattedString = """ **{}** * {} * * {} * * {} * **{}** """.format( "*" * 90, " " * 90, "{:<90}".format(reportHeading), " " * 90, "*" * 90, ) elif level == 3: # task level formattedString = """ --{}-- - {} - --{}-- """.format( "-" * 90, "{:<90}".format(reportHeading), "-" * 90, ) elif level == 4: # caption within task execution formattedString = """..{}.. . {} . ..{}..""".format( "." * 90, "{:<90}".format(reportHeading), "." * 90, ) if emit: self.info(formattedString, nl=newLine, tm=not continuation, force=True)Map the nodes of a node type between two versions of a TF dataset.
There are two scenarios in which you can use this object:
Extend a slot map to a node mapping.
If this object is initialized with a
slotMap, it can extend that mapping to a complete node mapping and save the mapping as anf"omap@{va}-{vb}"edge feature in the dataset versionvb.In this use case, the APIs for both
vaandvbshould be full TF APIs, in particular, they should provide thetf.core.localityAPI.Use an existing node map to upgrade features from one version to another.
In this use case, you do not have to pass a
slotMap, but in thevbdataset there should be anomap@a-bfeature, i.e. a node map from the nodes ofvato those ofvb.The APIs for both
vaandvbmay be partial TF APIs, which means that they only have to provide theNodeFeatures(F) andEdgeFeatures(E) APIs.Parameters
api:dict- TF API objects to several versions of a dataset, keyed by version label
va:string- version label of the version whose nodes are the source of the mapping
vb:string- version label of the version whose nodes are the target of the mapping
slotMap:dict, optionalNone- The actual mapping between slots of the old version and the new version
silent:string, optionalSILENT_D- See
Timestamp
Methods
def caption(self, level, heading, good=None, newLine=True, continuation=False)-
Expand source code Browse git
def caption(self, level, heading, good=None, newLine=True, continuation=False): silent = self.silent prefix = "" if good is None else "SUCCES " if good else "FAILURE " reportHeading = "{}{}".format(prefix, heading) emit = ( silent == VERBOSE or silent == AUTO and level in {-1, 2, 3, 4} or silent == TERSE and level in {2, 3} ) if level in {-1, 0}: # non-heading message decoration = "" if continuation else "| " formattedString = """{}{}""".format(decoration, reportHeading) elif level == 1: # pipeline level formattedString = """ ##{}## # {} # # {} # # {} # ##{}## """.format( "#" * 90, " " * 90, "{:<90}".format(reportHeading), " " * 90, "#" * 90, ) elif level == 2: # repo level formattedString = """ **{}** * {} * * {} * * {} * **{}** """.format( "*" * 90, " " * 90, "{:<90}".format(reportHeading), " " * 90, "*" * 90, ) elif level == 3: # task level formattedString = """ --{}-- - {} - --{}-- """.format( "-" * 90, "{:<90}".format(reportHeading), "-" * 90, ) elif level == 4: # caption within task execution formattedString = """..{}.. . {} . ..{}..""".format( "." * 90, "{:<90}".format(reportHeading), "." * 90, ) if emit: self.info(formattedString, nl=newLine, tm=not continuation, force=True) def exploreNodeMapping(self, nodeType)-
Expand source code Browse git
def exploreNodeMapping(self, nodeType): va = self.va vb = self.vb diagnosis = self.diagnosis self.caption(4, "Statistics for {} ==> {} ({})".format(va, vb, nodeType)) diag = diagnosis[nodeType] total = len(diag) if total == 0: return reasons = collections.Counter() for (n, dia) in diag.items(): reasons[dia] += 1 self.caption(-1, "\t{:<30} : {:6.2f}% {:>7}x".format("TOTAL", 100, total)) for stat in STAT_LABELS: statLabel = STAT_LABELS[stat] amount = reasons[stat] if amount == 0: continue perc = 100 * amount / total self.caption( -1, "\t{:<30} : {:6.2f}% {:>7}x".format(statLabel, perc, amount) ) def getDiagnosis(self, node=None, label=None)-
Expand source code Browse git
def getDiagnosis(self, node=None, label=None): """Show the diagnosis of a mapping. You can get various amounts of information about this: * A dictionary that maps all nodes of a given type to a diagnostic label * A list of nodes of a node type that have a given diagnostic label * The label of a given node Parameters ---------- node: string | integer, optional None The node or node type that you want to diagnose. If a string, it is a node type, and you get information about the nodes of that node type. If an int, it is a node, and you get information about that node. If `None`, you get information about all nodes. label: char, optional None If given, it is a diagnostic label (see `tf.dataset.nodemaps`), and the result is dependent on the value passed to *node*: if that is a single node, a boolean is returned telling whether that node has the given label; if that is a node type, a tuple is returned of all nodes in that node type that have this label; if that is None, a tuple is returned of all nodes with that label. If not given, the result depends again on the value passed to *node*: if that is a single node, you get its diagnostic label; if that is a node type, you get a dict, keyed by node, and valued by diagnostic label, for all nodes in the node type; if that is None, you get a dictionary keyed by node, and valued by diagnostic label but now for all nodes. """ diagnosis = self.diagnosis Fa = self.Fa otypesa = Fa.otype.s if label is None: return ( dict(chain.from_iterable(x.items() for x in diagnosis.values())) if node is None else diagnosis.get(node, {}) if type(node) is str else diagnosis.get(otypesa(node), {}).get(node, None) if type(node) is int else None ) else: return ( tuple( y[0] for y in chain.from_iterable(x.items() for x in diagnosis.values()) if y[1] == label ) if node is None else tuple( y[0] for y in diagnosis.get(node, {}).items() if y[1] == label ) if type(node) is str else diagnosis.get(otypesa(node), {}).get(node, None) == label if type(node) is int else None )Show the diagnosis of a mapping.
You can get various amounts of information about this:
- A dictionary that maps all nodes of a given type to a diagnostic label
- A list of nodes of a node type that have a given diagnostic label
- The label of a given node
Parameters
node:string | integer, optionalNone- The node or node type that you want to diagnose.
If a string, it is a node type, and you get information about the nodes
of that node type.
If an int, it is a node, and you get information about that node.
If
None, you get information about all nodes. label:char, optionalNone-
If given, it is a diagnostic label (see
tf.dataset.nodemaps), and the result is dependent on the value passed to node: if that is a single node, a boolean is returned telling whether that node has the given label; if that is a node type, a tuple is returned of all nodes in that node type that have this label; if that is None, a tuple is returned of all nodes with that label.If not given, the result depends again on the value passed to node: if that is a single node, you get its diagnostic label; if that is a node type, you get a dict, keyed by node, and valued by diagnostic label, for all nodes in the node type; if that is None, you get a dictionary keyed by node, and valued by diagnostic label but now for all nodes.
def legend(self)-
Expand source code Browse git
def legend(self): """Show the labels and descriptions of the diagnosis classes. When the mapping turns out to be not perfect for a node, the result can be categorized according to severity. This function shows that classification. See also `tf.dataset.nodemaps`. """ for (acro, desc) in STAT_LABELS.items(): self.info(f"{acro} = {desc}", tm=False)Show the labels and descriptions of the diagnosis classes.
When the mapping turns out to be not perfect for a node, the result can be categorized according to severity.
This function shows that classification. See also
tf.dataset.nodemaps. def makeNodeMapping(self, nodeType)-
Expand source code Browse git
def makeNodeMapping(self, nodeType): edge = self.edge if edge is None: self.error("Cannot make node mapping if no slot mapping is given") return False va = self.va vb = self.vb Lb = self.Lb Ea = self.Ea Eb = self.Eb Fa = self.Fa Lub = Lb.u Eosa = Ea.oslots.s Eosb = Eb.oslots.s otypesa = Fa.otype.s self.caption(2, f"Mapping {nodeType} nodes {va} ==> {vb}") diag = {} self.caption(0, f"Extending slot mapping {va} ==> {va} for {nodeType} nodes") for n in otypesa(nodeType): slots = Eosa(n) mappedSlots = set( chain.from_iterable(set(edge[s]) for s in slots if s in edge) ) mappedNodes = set( chain.from_iterable(set(Lub(s, nodeType)) for s in mappedSlots) ) result = {} nMs = len(mappedNodes) if nMs == 0: diag[n] = "a" elif nMs >= 1: theseMSlots = {} for m in mappedNodes: mSlots = set(Eosb(m)) dis = len(mappedSlots | mSlots) - len(mappedSlots & mSlots) result[m] = dis theseMSlots[m] = mSlots # we wait further case analysis before we put these counterparts of n into the edge if nMs == 1: m = list(mappedNodes)[0] dis = result[m] if dis == 0: diag[n] = "b" edge[n] = {m: None} # this is the most freqent case, hence an optimization: no dis value. # all other cases require the dis value to be passed on, even if 0 else: diag[n] = "c" edge[n] = {m: dis} else: edge[n] = result dis = min(result.values()) if dis == 0: diag[n] = "d" else: allMSlots = set( chain.from_iterable( set(theseMSlots[m]) for m in mappedNodes ) ) composed = allMSlots == mappedSlots and sum( result.values() ) == len(mappedSlots) * (len(mappedNodes) - 1) if composed: diag[n] = "f" else: diag[n] = "e" self.diagnosis[nodeType] = diag self.caption(0, "\tDone") def makeVersionMapping(self)-
Expand source code Browse git
def makeVersionMapping(self): Fa = self.Fa self.diagnosis = {} nodeTypes = Fa.otype.all for nodeType in nodeTypes[0:-1]: self.makeNodeMapping(nodeType) self.exploreNodeMapping(nodeType) self.writeMap() def migrateFeatures(self, featureNames, silent=None, location=None)-
Expand source code Browse git
def migrateFeatures(self, featureNames, silent=None, location=None): """Migrate features from one version to another based on a node map. If you have a dataset with several features, and if there is a node map between the versions, you can migrate features from the older version to the newer. The naive approach per feature is: For each node in the old version that has a value for the feature, map the node to the new version. That could be several nodes. Then give all those nodes the value found on the node in the old set. The problem with that is this: Suppose node `n` in the old version maps to nodes `p` and `q` in the new version, but not with the same quality. Suppose the value is `v`. Then the feature assignment will assign `v` to nodes `p` and `q`. But there could very well be a later node `m` with value `w` in the old version that also maps to `p`, and with a lower quality. Then later the node `p` in the new version will be assigned the value `w` because of this, and this is sub optimal. So we make sure that for each feature and each node in the new version the value assigned to it is the value of the node in the old version that maps with the highest quality to the new node. Parameters ---------- featureNames: tuple The names of the features to migrate. They may be node features or edge features or both. location: string, optional None If absent, the migrated features will be saved in the newer dataset. Otherwise it is a path where the new features should be saved. silent: string, optional None See `tf.core.timestamp.Timestamp` This will override the `silent` setting that has been used when creating the `Versions` object. """ silent = self.silent if silent is None else silentConvert(silent) omapName = self.omapName() TFa = self.TFa TFb = self.TFb TFb.setSilent(silent) TFb.warning("start migrating") TFb.load(omapName, add=True, silent=silent) Fsa = self.Fsa Esa = self.Esa Esb = self.Esb va = self.va vb = self.vb emap = Esb(omapName).f nodeFeatures = {} edgeFeatures = {} metaData = {} def getBestMatches(n, dest): result = [] mapped = emap(n) if mapped: perfect = [x for x in mapped if x[1] is None] if perfect: return [perfect[0]] nearPerfect = [x for x in mapped if x[1] == 0] if nearPerfect: (m, q) = nearPerfect[0] if m not in dest or m in dest and dest[m][1] is not None: return [nearPerfect[0]] for (m, q) in mapped: # these q cannot be None or 0 if m in dest: (origVal, origQ) = dest[m] higherQuality = origQ is not None and q < origQ if origVal == val: if higherQuality: result.append((m, q)) elif higherQuality: result.append((m, q)) else: result.append((m, q)) return result for featureName in featureNames: isEdge = TFa.features[featureName].isEdge TFb.info(f"Mapping {featureName} ({'edge' if isEdge else 'node'})") featureObj = (Esa if isEdge else Fsa)(featureName) featureInfo = dict(featureObj.items()) metaData[featureName] = featureObj.meta metaData[featureName]["upgraded"] = f"‼️ from version {va} to {vb}" edgeValues = featureObj.doValues if isEdge else False dest = edgeFeatures if isEdge else nodeFeatures dest[featureName] = {} dest = dest[featureName] # we will not only store the feature value in dest, # but also the quality of the mapping that # produced the value. # We only override an existing value # if the quality of the mapping is better. # Moreover, if one mapping has quality 0, # it means that the mapping is perfect regarding the material in both nodes # but there are lesser mappings as well. # In those cases we do not extend the value to those lesser mappings # Later, we strip the qualities again. if isEdge: if edgeValues: for (n, ts) in featureInfo.items(): matched = getBestMatches(n, dest) if matched: matchedTs = {} for (t, val) in ts.items(): for (t, r) in getBestMatches(t, matchedTs): matchedTs[t] = (val, r) for t in matchedTs: matchedTs[t] = matchedTs[t][0] for (m, q) in matched: dest[m] = (matchedTs, q) """ mapped = emap(n) if mapped: mappedts = {} for (t, val) in ts.items(): tmapped = emap(t) if tmapped: for tEntry in tmapped: mappedts[tEntry[0]] = val for entry in mapped: dest[entry[0]] = mappedts """ else: for (n, ts) in featureInfo.items(): matched = getBestMatches(n, dest) if matched: matchedTs = {} for t in ts: for (t, r) in getBestMatches(t, matchedTs): matchedTs[t] = (None, r) matchedTs = set(matchedTs) for (m, q) in matched: dest[m] = (matchedTs, q) """ mapped = emap(n) if mapped: mappedts = set() for t in ts: tmapped = emap(t) if tmapped: for tEntry in tmapped: mappedts.add(tEntry[0]) for entry in mapped: dest[entry[0]] = mappedts """ else: for (n, val) in featureInfo.items(): for (m, q) in getBestMatches(n, dest): dest[m] = (val, q) for m in dest: dest[m] = dest[m][0] TFb.save( nodeFeatures=nodeFeatures, edgeFeatures=edgeFeatures, metaData=metaData, location=location, module=vb, silent=silent, ) TFb.warning("Done")Migrate features from one version to another based on a node map.
If you have a dataset with several features, and if there is a node map between the versions, you can migrate features from the older version to the newer.
The naive approach per feature is:
For each node in the old version that has a value for the feature, map the node to the new version. That could be several nodes. Then give all those nodes the value found on the node in the old set.
The problem with that is this:
Suppose node
nin the old version maps to nodespandqin the new version, but not with the same quality. Suppose the value isv. Then the feature assignment will assignvto nodespandq.But there could very well be a later node
mwith valuewin the old version that also maps top, and with a lower quality. Then later the nodepin the new version will be assigned the valuewbecause of this, and this is sub optimal.So we make sure that for each feature and each node in the new version the value assigned to it is the value of the node in the old version that maps with the highest quality to the new node.
Parameters
featureNames:tuple- The names of the features to migrate. They may be node features or edge features or both.
location:string, optionalNone- If absent, the migrated features will be saved in the newer dataset. Otherwise it is a path where the new features should be saved.
silent:string, optionalNone- See
TimestampThis will override thesilentsetting that has been used when creating theVersionsobject.
def omapName(self)-
Expand source code Browse git
def omapName(self): va = self.va vb = self.vb return f"omap@{va}-{vb}" def writeMap(self)-
Expand source code Browse git
def writeMap(self): TFb = self.TFb va = self.va vb = self.vb edge = self.edge silent = self.silent fName = self.omapName() self.caption(4, "Write edge as TF feature {}".format(fName)) edgeFeatures = {fName: edge} metaData = { fName: { "about": "Mapping from the slots of version {} to version {}".format( va, vb ), "encoder": "TF tf.dataset.nodemaps", "valueType": "int", "edgeValues": True, } } TFb.save( nodeFeatures={}, edgeFeatures=edgeFeatures, metaData=metaData, silent=silent, )